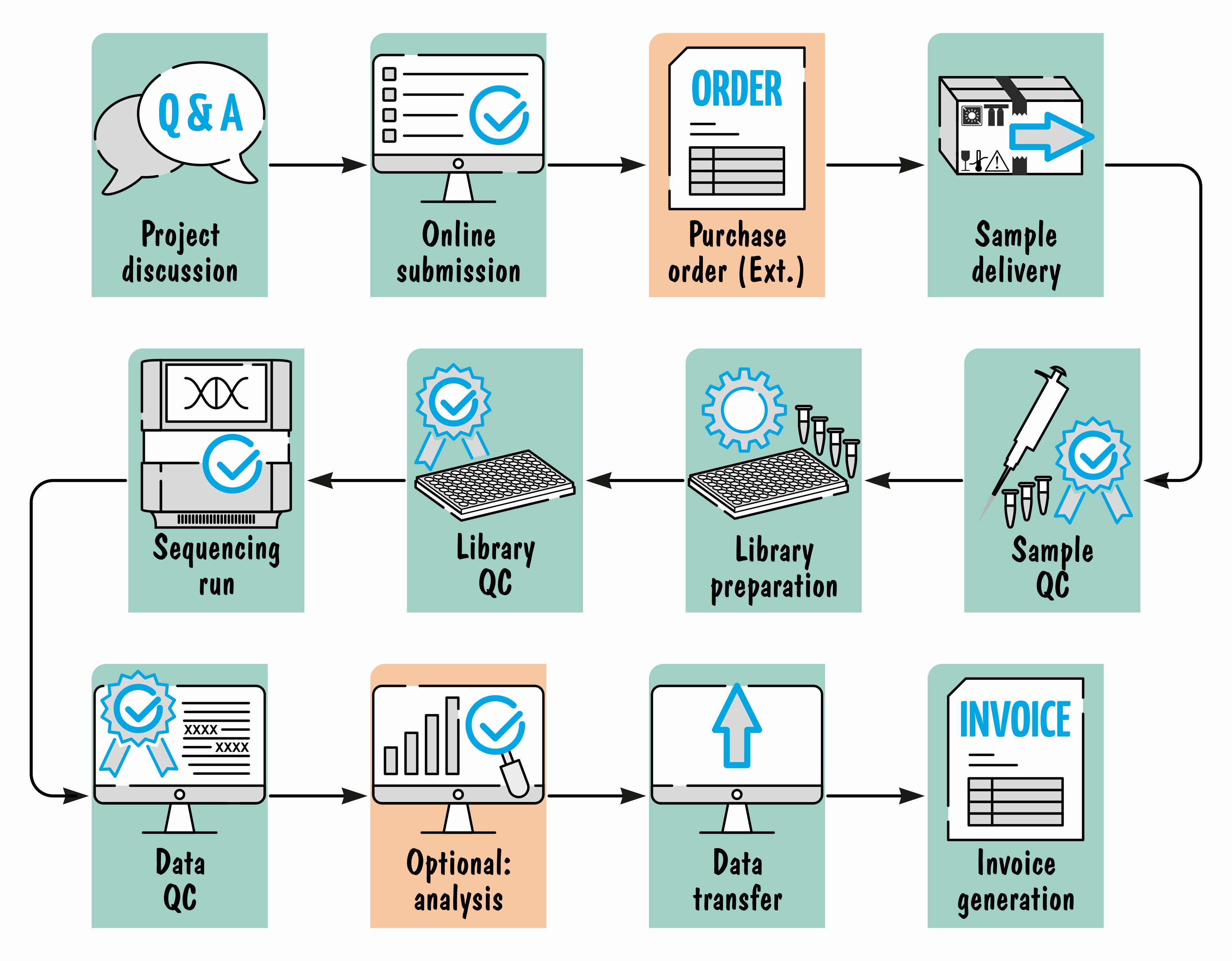
Service workflow#
Sequencing projects#
We offer a broad range of services and to get a personalized quote, please contact us with the following details:
- Number of samples
- Sequencing depth (expected Gb of data per sample) needed
- Expected time-frame of availability of samples
- User and PI affiliation (institution)
Short read sequencing#
Genomics
- Whole genome sequencing (small genomes)
- Metagenomics/ shotgun sequencing
- Chromatin immunoprecipitation sequencing (ChIP-Seq)
- Assay for Transposase-Accessible Chromatin Sequencing (ATAC-Seq)
- DNase I hypersensitive sites sequencing (DNase I-Seq)
- Reduced representation bisulfite sequencing (RRBS)
- Methylated DNA immumoprecipitation sequencing (MeDIP-Seq)
- Exome sequencing
- Immuno-profiling (TCR & BCR repertoire)
Transcriptomics
- totalRNA Stranded sequencing (rRNA depletion)
- mRNA Stranded sequencing (mRNA-Seq)
- 3’ RNA sequencing
- Low input RNA sequencing
- Meta transcriptome sequencing
- Small RNA sequencing (smRNA-Seq)
Targeted Resequencing
- RNA (gene panel)
- DNA (gene panel)
- Amplicon sequencing
Long read sequencing#
- Metagenome sequencing
- Whole genomic sequencing
- mtDNA sequencing
- amplicon sequencing
- full length 16S/18S rRNA gene sequencing
- full length cDNA sequencing
- direct RNA sequencing
In case you have a project with an application not listed above, please contact us regarding the feasibility of your project.
Single-cell projects#
For all the single cell projects we suggest having a kickoff meeting before starting the project. No single cell project will be entertained without prior discussion of time of reception of samples and experimental strategies.
Droplet based methods#
DropSeq
- Single cell RNA sequencing (scRNA-Seq)
10X Genomics
- Single cell RNA sequencing (scRNA-Seq)
- Single cell fixed RNA sequencing
- Single nuclear RNA sequencing (snRNA-Seq)
- Single cell ATAC sequencing (scATAC-Seq)
- Single cell multiome sequencing (scATAC and scRNA-seq)
- Cellular indexing of transcriptomes and epitopes by sequencing (CITE-Seq)
- Single cell V(D)J sequencing
Cell sorting based method#
- Single cell RNA sequencing (scRNA-Seq)
single cell sorted in 96well plate format. For this method please collect the PCR plates with the lysis buffer from the platform prior to cell sorting.
Microwell based method#
- Single cell RNA sequencing (scRNA-Seq)
- Single nuclear RNA sequencing (snRNA-Seq)
- Single cell glycosylation sequencing
- Single cell V(D)J sequencing
In case you have a project with an application not listed above, please contact us regarding the feasibility of your project.
Data analysis#
LCSB genomics platform offers the primary data analysis and provide the following files to the user.
- Fastq files after demultiplexing (Illumina) OR fast5 without de-multiplexing (Oxford Nanopore)
- Quality control report from fastQC software (Illumina).
In addition to the above primary data analysis for some cases the platform provides the preliminary analysis results upon request
- Gene expression using DESeq using Dragen BioIT server
- Single cell gene expression matrix (10X Genomics) using Dragen BioIT server
- Single cell gene expression matrix (DropSeq) using in-house bash scripts
For advanced secondary analysis we can recommend contacting PIs from bioinformatics core. But these services are separate to that of LCSB genomics platform and therefore, the conditions need to be inquired independently by the user.